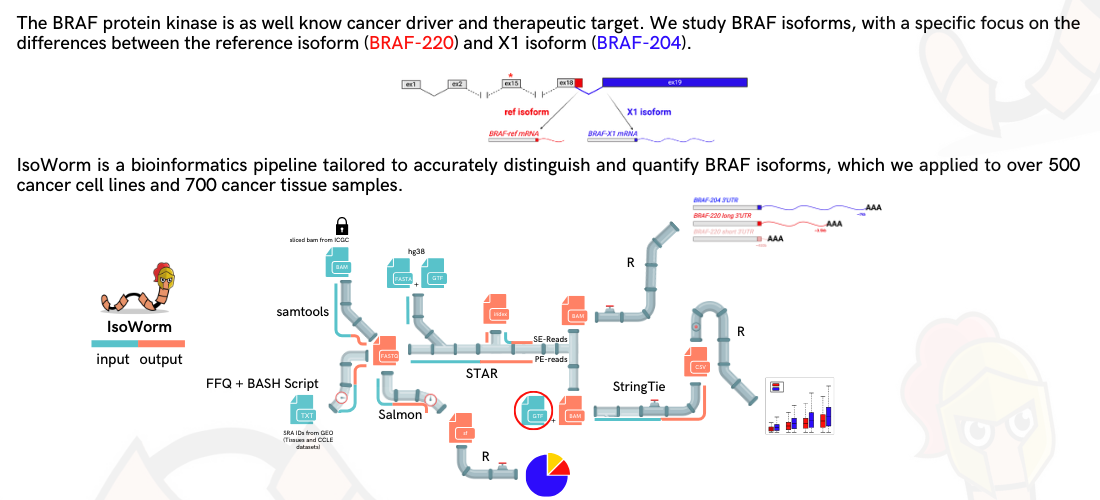
The BRAF protein kinase is extensively researched due to its role as a cancer driver and potential therapeutic target. Our study focuses on quantifying and analyzing clinical outcomes related to BRAF isoforms, specifically examining the ratio between the primary isoform (BRAF-220) and an alternative one (BRAF-204). We developed IsoWorm, a bioinformatics pipeline tailored to accurately distinguish and quantify BRAF isoforms, which we applied to over 500 cancer cell lines and 700 cancer tissue samples. Thanks to FLIBase database, we also re-analyzed TCGA data on more than 9000 cancer tissue samples. We consistently found that BRAF-204 is the most abundant BRAF isoform in human cancer and it is 1.5 to 75 times more expressed than BRAF-220. Crucially, we identified colon and kidney as cancer types where extreme BRAF-204/BRAF-220 ratios are associated with different outcomes, in terms of overall survival and progression-free interval. Together with the experimental characterization undertaken over the years, our in silico analyses ultimately establish BRAF as a mix of isoforms, with BRAF-204 being much more expressed than BRAF-220. These findings prompt the undertaking of a systematic benchmarking of the isoforms in terms of molecular mechanisms, biological activities and clinical relevance.